Constraint and Contingency in Multifunctional Gene Regulatory Circuits
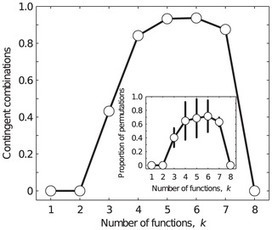
Many essential biological processes, ranging from embryonic patterning to circadian rhythms, are driven by gene regulatory circuits, which comprise small sets of genes that turn each other on or off to form a distinct pattern of gene expression. Gene regulatory circuits often have multiple functions. This means that they can form different gene expression patterns at different times or in different tissues. We know little about multifunctional gene regulatory circuits. For example, we do not know how multifunctionality constrains the evolution of such circuits, how many circuits exist that have a given number of functions, and whether tradeoffs exist between multifunctionality and the robustness of a circuit to mutation. Because it is not currently possible to answer these questions experimentally, we use a computational model to exhaustively enumerate millions of regulatory circuits and all their possible functions, thereby providing the first comprehensive study of multifunctionality in model regulatory circuits. Our results highlight limits of circuit designability that are relevant to both systems biologists and synthetic biologists.
Payne JL, Wagner A (2013) Constraint and Contingency in Multifunctional Gene Regulatory Circuits. PLoS Comput Biol 9(6): e1003071.http://dx.doi.org/10.1371/journal.pcbi.1003071
No hay comentarios:
Publicar un comentario